Abstract
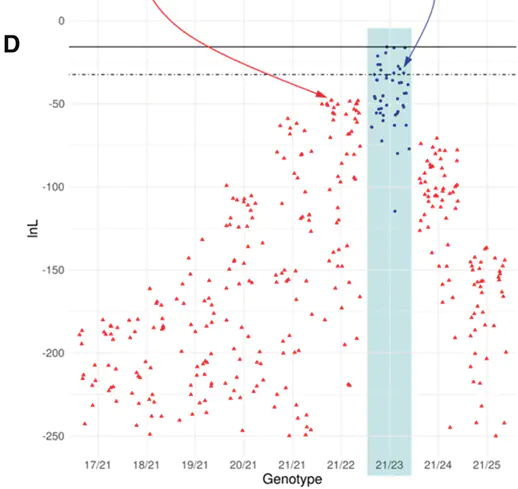
Motivation Massively parallel capture of short tandem repeats (STRs, or microsatellites) provides a strategy for population genomic and demographic analyses at high resolution with or without a reference genome. However, the high Polymerase Chain Reaction (PCR) cycle numbers needed for target capture experiments create genotyping noise through polymerase slippage known as PCR stutter. Results We developed SONiCS—Stutter mONte Carlo Simulation—a solution for stutter correction based on dense forward simulations of PCR and capture experimental conditions. To test SONiCS, we genotyped a 2499-marker STR panel in 22 humpback dolphins (Sousa sahulensis) using target capture, and generated capillary-based genotypes to validate five of these markers. In these 110 comparisons, SONiCS showed a 99.1% accuracy rate and a 98.2% genotyping success rate, miscalling a single allele in a marker with low sequence coverage and rejecting another as un-callable. Availability and implementation Source code and documentation for SONiCS is freely available on Github. Raw read data used in experimental validation of SONiCS have been deposited in the Sequence Read Archive under accession number SRP135756.