Genomic analysis of DNA repair genes and androgen signaling in prostate cancer
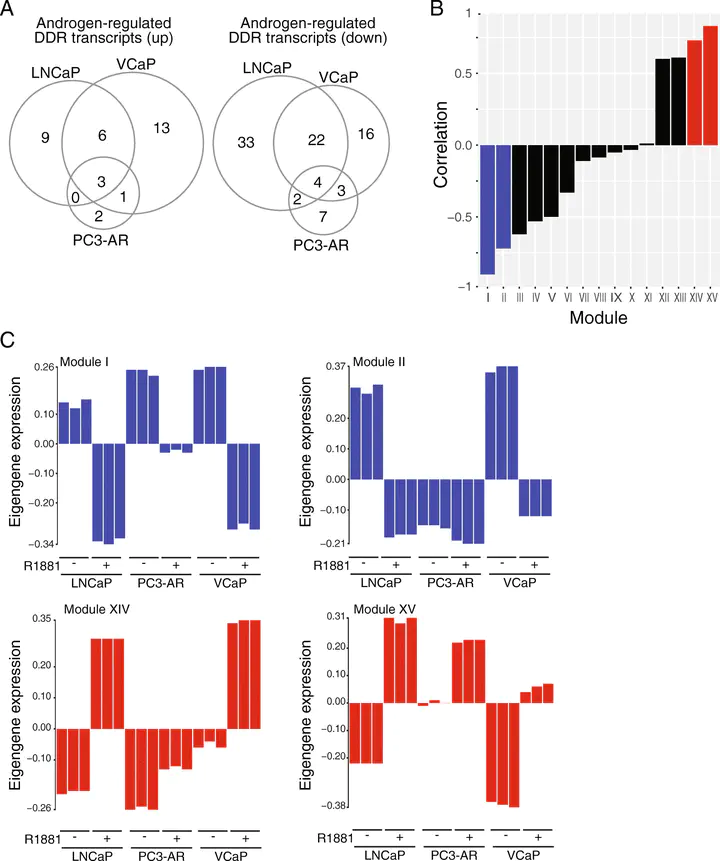 Comparison of DNA damage response gene expression in prostate cancer cell lines.
Comparison of DNA damage response gene expression in prostate cancer cell lines.Abstract
Background The cellular effects of androgen are transduced through the androgen receptor, which controls the expression of genes that regulate biosynthetic processes, cell growth, and metabolism. Androgen signaling also impacts DNA damage signaling through mechanisms involving gene expression and transcription-associated DNA damaging events. Defining the contributions of androgen signaling to DNA repair is important for understanding androgen receptor function, and it also has translational implications. Methods We generated RNA-seq data from multiple prostate cancer lines and used bioinformatic analyses to characterize androgen-regulated gene expression. We compared the results from cell lines with gene expression data from prostate cancer xenografts, and patient samples, to query how androgen signaling and prostate cancer progression influences the expression of DNA repair genes. We performed whole genome sequencing to help characterize the status of the DNA repair machinery in widely used prostate cancer lines. Finally, we tested a DNA repair enzyme inhibitor for effects on androgen-dependent transcription. Results Our data indicates that androgen signaling regulates a subset of DNA repair genes that are largely specific to the respective model system and disease state. We identified deleterious mutations in the DNA repair genes RAD50 and CHEK2. We found that inhibition of the DNA repair enzyme MRE11 with the small molecule mirin inhibits androgen-dependent transcription and growth of prostate cancer cells. Conclusions Our data supports the view that crosstalk between androgen signaling and DNA repair occurs at multiple levels, and that DNA repair enzymes in addition to PARPs, could be actionable targets in prostate cancer.