COVID-19 mortality in England and Wales
Based on the ONS data
Disclaimer
As probably many of the data scientist, I wanted to ease my anxiety caused by the COVID-19. I decided to look at the data related to the pandemic. I decided not to look at the daily cases as John Burn-Murdoch from Financial Times and the team Our World in Data are doing a great job. The reasons why I decided to look at deaths are explained in the Motivation section.
I am a computational biologist and all the plots I am making here are just observations. I am not claiming any conclusions, nor do I do any predictions. All the code is open source, and free for anyone to use, available on github.
Also, very warm thanks to Kaspar for his help with decisions regarding color schemes and his helpful advice.
Motivation
I decided to look into the mortality rates because I wanted to put the numbers reported by the government in some context. My first question was whether the deaths reported as COVID-19 deaths were a subset of deaths one could expect to see this year? How much COVID-19 increases the death toll? And finally, how does the age distribution of fatalities look?
The data I plotted below shades a bit of light on those topics, but for a more in-depth answer, I am afraid we will have to wait for long. Nevertheless, I still think the following is quite interesting. If not for others, then for the sole reason of looking at the data in almost real-time.
Setup
Loading up the necessary packages (installing them if they are missing). Setting up defaults and tweaking the plotting defaults.
# Load packages
cran_pkgs <- c("tidyverse", "ggsci", "patchwork",
"readxl", "knitr", "ggforce", "lubridate")
if (grepl("content", getwd())) {setwd(paste0(sub(pattern = "(.*/content)/.*",
replacement = "\\1",
x = getwd()), "/post/covid-19"))}
source("scripts/load_pkgs.R")
load_packages(cran_pkgs = cran_pkgs)
# Set defaults for plotting
theme_set(theme_bw() + theme(legend.position = "bottom"))
# Don't print warnings
knitr::opts_chunk$set(warning = FALSE, message = FALSE)
# Update date of the data
data_up_to <- ymd("2020-05-22")
Data
Data up to: r ymd(data_up_to)
Data is coming from Office for National Statistics, Deaths registered weekly in England and Wales, provisional dataset. For now I am downloading the data manually into the data directory. It would be great to download it directly.
Weekly figures
Both 2020 and 2019 figures for comparison of the trend. The 2020 will be changed each week, while 2019 remains stable.
Note: Unfortunately due to formatting of the data the following code is not entirely reproducible as there are parts of it that would have to be adjusted with each newiteration of the spreadsheet.
df_2019 <-
read_xls("./data/publishedweek522019.xls",
sheet = "Weekly figures 2019", skip = 3)[c(4,5,9),-c(1,2)] %>%
mutate(value = c("total_deaths", "total_deaths_ave5", "respiratory_deaths")) %>%
mutate_at(vars(-value), as.numeric) %>%
pivot_longer(names_to = "week",
values_to = "number_of_deaths", -value) %>%
mutate(year = "2019")
df_2015 <-
read_xls("./data/publishedweek2015.xls",
sheet = "Weekly Figures 2015", skip = 3)[c(4,5,9),-c(1)] %>%
mutate(value = c("total_deaths", "total_deaths_ave5", "respiratory_deaths")) %>%
mutate_at(vars(-value), as.integer) %>%
pivot_longer(names_to = "week",
values_to = "number_of_deaths", -value) %>%
mutate(year = "2015",
week = as.integer(week),
week_ = ymd("2020-01-03") + 7 * (week-1))
current_2020_spreadsheet <- "./data/publishedweek212020.xlsx"
sheets_2020 <- readxl::excel_sheets(current_2020_spreadsheet)
df_2020_ <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[5], skip = 4)[c(4,6,13,14),-c(1,2)] %>%
mutate(value2 = c("total_deaths", "total_deaths_ave5",
"respiratory_deaths", "covid_deaths"),
value = c("total_deaths", "total_deaths_ave5",
"respiratory_deaths", "respiratory_deaths")) %>%
mutate_at(vars(-value, -value2), as.numeric) %>%
pivot_longer(names_to = "week",
values_to = "number_of_deaths",
-c(value, value2))
df_2020 <-
df_2020_ %>%
select(-value2) %>%
mutate(year = "2020") %>%
filter(!is.na(number_of_deaths))
df_all <- bind_rows(list(df_2019, df_2020)) %>%
mutate(week = as.numeric(week),
week_ = ymd("2020-01-03") + 7 * (week-1)) %>%
group_by(value, week, week_, year) %>%
summarise(number_of_deaths = sum(number_of_deaths)) %>%
ungroup()
COVID-19 specific datasets
place_of_death <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[grepl("Place", sheets_2020)],
skip = 4)[14:20,1:7] %>%
rename(place = 1) %>%
pivot_longer(names_to = "country",
values_to = "number_of_deaths",
c(-place)) %>%
mutate(country = rep(c(rep("England and Wales", 2), rep("England", 2), rep("Wales", 2)), 7),
cause = rep(rep(c("Total deaths", "COVID-19 deaths"), 3), 7)) %>%
mutate(number_of_deaths = as.numeric(number_of_deaths))
actual_vs_reported <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[str_detect(sheets_2020, regex("E&W comparisons", ignore_case = TRUE))], skip = 4,
col_types = c("date", rep("numeric", 8)))[, 1:9] %>%
filter(!is.na(Date)) %>%
pivot_longer(names_to = "source",
values_to = "number_of_deaths",
-Date)
place_of_death_long <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[grepl("Place", sheets_2020)],
skip = 4)[4:9,] %>%
rename(place = 1) %>%
pivot_longer(names_to = "cause",
values_to = "number_of_deaths",
-place) %>%
mutate(cause = rep(c("Total deaths", "COVID-19 deaths"), n()/2),
country = rep(c(rep("England and Wales", 2),
rep("England", 2),
rep("Wales", 2)), n()/6),
week = rep(rep(11:(10 + n() / 36), each = 6), 6))
age_distribution <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[str_detect(sheets_2020,
regex("covid-19 .* occurrences",
ignore_case = TRUE))],
skip = 4)[c(7:26,29:48,51:70), -1] %>%
mutate(sex = c(rep("Both", 20), rep("Males", 20), rep("Females", 20))) %>%
rename(age_group = `...2`) %>%
mutate_at(vars(-age_group, -sex), as.numeric) %>%
pivot_longer(names_to = "week",
values_to = "number_of_deaths",
cols = -c(age_group, sex)) %>%
replace_na(list(number_of_deaths = 0)) %>%
group_by(sex, week) %>%
filter(sum(number_of_deaths, na.rm = TRUE) > 0)
age_levels <- age_distribution$age_group %>% unique()
age_distribution <-
age_distribution %>%
ungroup() %>%
mutate(week = as.numeric(week),
week_ = lubridate::ymd("2020-01-03") + 7 * (week-1)) %>%
mutate(age_group = factor(age_group, levels = age_levels))
age_distribution_all <-
read_xlsx(current_2020_spreadsheet,
sheet = sheets_2020[str_detect(sheets_2020,
regex("Weekly figures 2020",
ignore_case = TRUE))],
skip = 4)[c(17:36,39:58,61:80), -1] %>%
mutate(sex = c(rep("Both", 20), rep("Males", 20), rep("Females", 20))) %>%
rename(age_group = 1) %>%
mutate_at(vars(-age_group, -sex), as.numeric) %>%
pivot_longer(names_to = "week",
values_to = "number_of_deaths",
cols = -c(age_group, sex)) %>%
replace_na(list(number_of_deaths = 0)) %>%
group_by(sex, week) %>%
filter(sum(number_of_deaths, na.rm = TRUE) > 0) %>%
ungroup() %>%
mutate(week = as.numeric(week),
week_ = lubridate::ymd("2020-01-03") + 7 * (week-1)) %>%
mutate(age_group = factor(age_group, levels = age_levels))
Mortality in the context of past years
All weekly deaths in England and Wales
Still very early, but looks like the overall weekly numbers are higher than last year, and the past 5 years.
total <-
df_all %>%
filter(value == "total_deaths") %>%
ggplot(aes(x = week_, y = number_of_deaths, fill = year)) +
geom_bar(stat = "identity", position = "dodge") +
geom_smooth(data = filter(df_all, value == "total_deaths_ave5" & year == "2019"),
color = "grey60", se = FALSE) +
facet_zoom(xy = week_ < data_up_to + 1,
horizontal = FALSE, show.area = FALSE) +
labs(y = "Number of deaths",
title = "Weekly total deaths [grey = 5 year average]") +
theme(axis.title.x = element_blank()) +
scale_fill_aaas()
total
Respiratory deaths
COVID-19 deaths are increasing the number of repiratory deaths.
upper_limit <-
df_all %>%
filter(value == "respiratory_deaths") %>%
pull(number_of_deaths) %>%
max() + 100
respiratory <-
df_all %>%
filter(value == "respiratory_deaths") %>%
ggplot(aes(x = week_, y = number_of_deaths,
group = year, color = year, shape = year)) +
geom_point() +
geom_smooth(se = FALSE, span = 0.3) +
facet_zoom(xy = week_ < data_up_to + 1,
horizontal = FALSE, show.area = FALSE) +
scale_color_aaas() +
labs(y = "Number of deaths", title = "Weekly respiratory deaths") +
theme(axis.title.x = element_blank()) +
coord_cartesian(ylim = c(0, upper_limit))
respiratory
However, the difference is not yet visible on the cummulative scale.
cumulative_respiratory <-
df_all %>%
arrange(week) %>%
group_by(value, year) %>%
mutate(cum = cumsum(number_of_deaths)) %>%
filter(value == "respiratory_deaths") %>%
ungroup() %>%
ggplot(aes(x = week_, y = cum, color = year, shape = year)) +
geom_point() +
geom_smooth(se = FALSE) +
facet_zoom(xy = week_ < data_up_to + 1,
horizontal = FALSE, show.area = FALSE) +
scale_color_aaas() +
labs(y = "Number of deaths", shape = "Year",
color = "Year", title = "Cumulative respiratory deaths") +
theme(axis.title.x = element_blank())
cumulative_respiratory
COVID-19 deaths increase exponentailly, while the other respiratory deths seem to be stable.
As per BBC plot I added the 2015 respiratory deaths, when the worst flu outbreak took place.
df_2020_ %>%
mutate(year = "2020",
value2 = gsub("respiratory_deaths", "other respiratory deaths", value2),
value2 = str_replace(value2, "covid_deaths", "COVID-19 deaths"),
week = as.numeric(week),
week_ = lubridate::ymd("2020-01-03") + 7 * (week-1)) %>%
bind_rows((df_2015 %>%
mutate(value2 = ifelse(value == "respiratory_deaths",
"respiratory deaths in 2015", value)))) %>%
filter(value == "respiratory_deaths") %>%
filter(!is.na(number_of_deaths),
week_ < data_up_to + 1) %>%
ggplot(aes(x = week_, y = number_of_deaths, color = value2, shape = value2)) +
geom_point() +
geom_smooth(se = FALSE, span = 0.3) +
scale_color_uchicago() +
labs(y = "Number of deaths", title = "Weekly respiratory deaths",
color = "Cause of death", shape = "Cause of death") +
theme(axis.title.x = element_blank(), legend.position = "bottom")
total / respiratory / cumulative_respiratory + plot_layout(guides = "collect")
ggsave("./plots/up_to200327.png")
COVID-19 specific stats
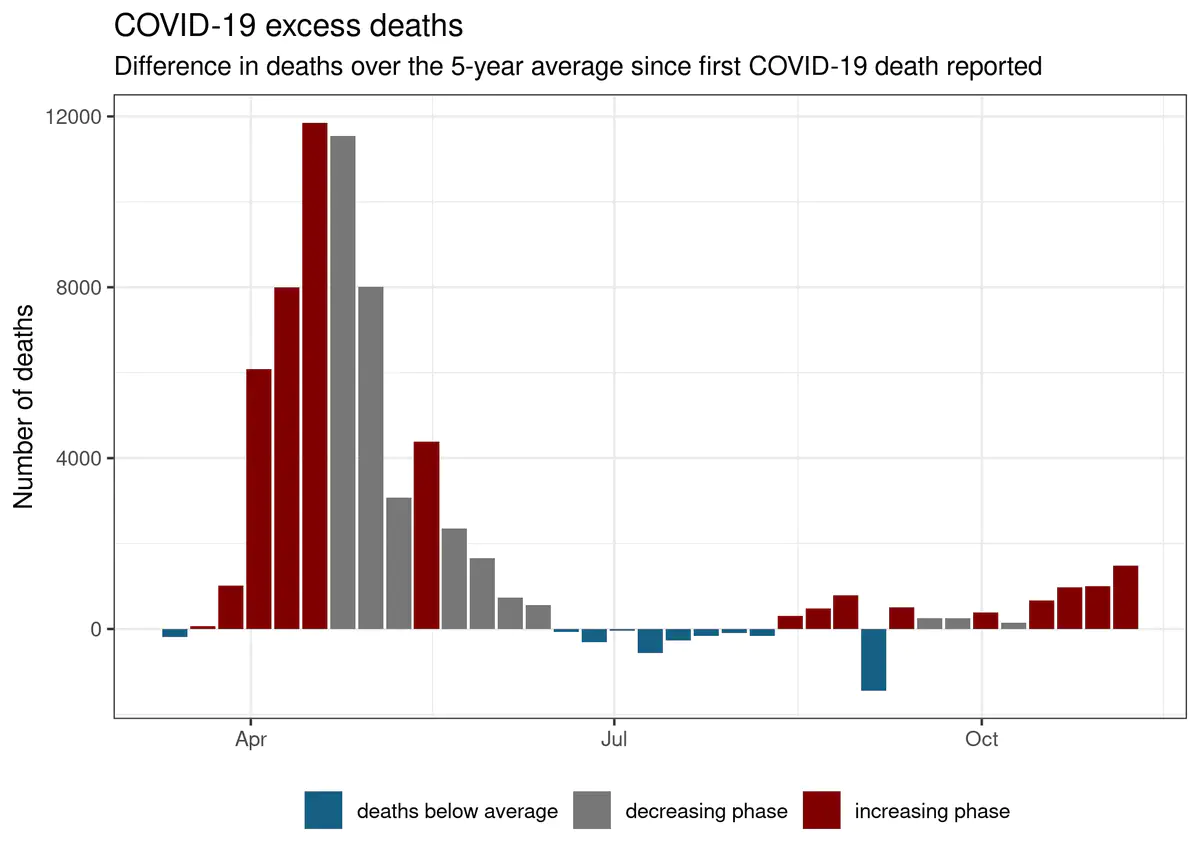
Excess deaths - proxy for all COVID-19 related deaths
In this plot, I am looking at the difference in total number of deaths reported in the period since the first COVID-19 related death was reported and the 5-year average.
col_scheme <- pal_uchicago()(5)[c(1:2,5)]
names(col_scheme) <- c("increasing phase",
"decreasing phase",
"deaths below average")
df_2020_excess <-
df_2020_ %>%
filter(!is.na(number_of_deaths)) %>%
group_by(week) %>%
filter(any((value2 == "covid_deaths") & (number_of_deaths > 0))) %>%
ungroup() %>%
filter(value2 %in% c("total_deaths", "total_deaths_ave5")) %>%
select(-value) %>%
pivot_wider(names_from = value2,
values_from = number_of_deaths) %>%
mutate(difference = total_deaths - total_deaths_ave5,
fract = difference / total_deaths_ave5,
week_ = lubridate::ymd("2020-01-03") + 7 * (as.numeric(week) - 1),
group = ifelse(difference < 0,
"deaths below average",
ifelse(difference > lag(difference),
"increasing phase",
"decreasing phase")))
all_excess_deaths <- df_2020_excess %>%
pull(difference) %>%
sum()
all_deaths <- df_2020 %>%
filter(value == "total_deaths") %>%
pull(number_of_deaths) %>%
sum()
fraction_excess <- all_excess_deaths / all_deaths
df_2020_excess %>%
ggplot(aes(week_, difference, fill = group)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = col_scheme) +
theme(legend.title = element_blank(), axis.title.x = element_blank()) +
labs(y = "Number of deaths",
title = "COVID-19 excess deaths",
subtitle = "Difference in deaths over the 5-year average since first COVID-19 death reported")
We see more evidence that England & Wales has moved past the peak. Up to r format(data_up_to, '%b, %d') at least r format(all_excess_deaths, big.mark =" ") more people died when compared to the 5 year average. This means that r sprintf("%.1f%%", 100 * fraction_excess) of all deaths this year were directly, or indirectly caused by COVID-19 (taking the excess mortality over the 5 year average as best proxy for number of deaths as a result of pandemic).
Place of death
fract_place_of_death <-
place_of_death %>%
filter(place != "All deaths",
country == "England and Wales") %>%
mutate(place = gsub("Hospital \\(acute or community, not psychiatric\\)",
"Hospital", place),
place = gsub("Other communal establishment",
"Other communal\nestablishment", place)) %>%
group_by(cause) %>%
mutate(fract = round(number_of_deaths / sum(number_of_deaths) * 100, 2))
As of r format(data_up_to, '%b, %d'), majority of the deaths are reported in hospitals. However r 100 - round((fract_place_of_death %>% filter(place == "Hospital", cause == "COVID-19 deaths") %>% pull(fract)), 0)% of deaths occur outside of hospitals. Thankfully, all numbers seem to be going down, with hospital deaths dropping below the numbers from the begining of the epidemic.
fract_place_of_death %>%
group_by(place) %>%
filter(max(fract) > 1) %>%
ggplot(aes(place, fract, fill = cause)) +
geom_bar(stat = "identity", position = "dodge") +
scale_fill_nejm() +
labs(title = paste0("Place of death [data up to ", data_up_to, "]"),
y = "Fraction of reported deaths",
fill = "Cause of death:") +
theme(axis.title.x = element_blank(), text = element_text(size = 16))
place_of_death_long %>%
filter(country == "England and Wales") %>%
mutate(place = gsub("Hospital \\(acute or community, not psychiatric\\)",
"Hospital", place),
place = gsub("Other communal establishment",
"Other communal\nestablishment", place),
week_ = ymd("2020-01-03") + 7 * (week-1),
number_of_deaths = as.numeric(number_of_deaths)) %>%
filter(place %in% c("Care Home", "Home", "Hospice", "Hospital")) %>%
ggplot(aes(week_, number_of_deaths, fill = place)) +
geom_bar(stat = "identity", position = "dodge") +
scale_fill_lancet() +
facet_grid(cause~place) +
labs(title = paste0("Place of death [data up to ", data_up_to, "]"),
y = "Number of reported deaths",
fill = "Place of death:") +
theme(axis.title.x = element_blank(),
text = element_text(size = 16)) +
scale_x_date(breaks = ymd("2020/03/13", "2020/04/03"), date_labels = "%b")
We saw increase in almost all places of death followed, thankfully by decrease.
My observations:
- COVID-19 accounted only for some of the increase.
My speculation: Was that the case because no test was carried out? I find it unlikely, as the deaths reported to ONS don’t have to be verified by testing to be classified as COVID-19 related. It might be, for example, because people are scared to go to the hospitals.
Also, as have been already reported, the outbreak in care homes started later, hence we are seeing an increase in COVID-19 related deaths in those facilities.
- We see that number of deaths with no mention of COVID-19 decreased in the hospitals and increased in recent weeks. I hypothesize it potentially can be explained by increase in deaths in other places, less road accidents and less scheduled medical interventions. And now, that lockdown has been lifted we potentially see the increase in accidents.
Note: Many thanks to German (and David Spiegelhalter plots) for making me realise a mistake in the earlier iteration of the following plot!
place_of_death_long %>%
filter(country == "England and Wales") %>%
mutate(place = gsub("Hospital \\(acute or community, not psychiatric\\)",
"Hospital", place),
place = gsub("Other communal establishment",
"Other communal\nestablishment", place),
week_ = ymd("2020-01-03") + 7 * (week-1),
number_of_deaths = as.numeric(number_of_deaths)) %>%
pivot_wider(names_from = "cause", values_from = number_of_deaths) %>%
mutate(`No COVID-19 mentioned` = `Total deaths` - `COVID-19 deaths`) %>%
select(-`Total deaths`) %>%
pivot_longer(names_to = "cause", values_to = "number_of_deaths",
-c(place, country, week, week_)) %>%
group_by(place) %>%
mutate(max_deaths = max(number_of_deaths)) %>%
ungroup() %>%
mutate(place = reorder(place, -max_deaths)) %>%
filter(place %in% c("Care Home", "Home", "Hospital")) %>%
ggplot(aes(week_, number_of_deaths, fill = cause)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = pal_nejm()(6)[c(1,6)]) +
facet_wrap(~place) +
labs(title = paste0("Place of death [data up to ", data_up_to, "]"),
y = "Number of reported deaths",
fill = "Place of death:") +
theme(axis.title.x = element_blank(),
text = element_text(size = 16)) +
scale_x_date(breaks = ymd("2020/03/13", "2020/04/03"), date_labels = "%b")
Differences between the reported numbers
The numbers reported by the government are lower than the actual number of deaths each day. Some of that comes from the delay in confirming the cause of death.
All the comparisons between previous years (in the earlier parts of this notebook) are done based on figures from ONS deaths by date of registration – registered by r format(data_up_to, '%d %b') set for compatibility with earlier datasets.
col_scheme <- c("#E18727", "#849460", "#56603F", "#20854E")
names(col_scheme) <- unique(actual_vs_reported$source)[1:4]
label_postions <-
actual_vs_reported %>%
filter(source %in% names(col_scheme)) %>%
group_by(source) %>%
mutate(max = max(number_of_deaths)) %>%
arrange(Date) %>%
mutate(deaths_on_that_day = number_of_deaths - lag(number_of_deaths),
max_on_day = max(deaths_on_that_day, na.rm = TRUE)) %>%
ungroup() %>%
filter(Date == lubridate::ymd("2020/03/6")) %>%
arrange(-max) %>%
mutate(fixed_position_cum = max(max) - 0.07 * max(max) * 1:length(col_scheme),
fixed_position_daily = max(max_on_day) - 0.07 * max(max_on_day) * 1:length(col_scheme))
discrepancies <-
actual_vs_reported %>%
filter(source %in% names(col_scheme)) %>%
ggplot(aes(x = Date, y = number_of_deaths, color = source, shape = source)) +
geom_smooth(se = FALSE) +
geom_point(color = "black") +
geom_text(data = label_postions, size = 7, hjust = 0,
aes(x = Date, y = fixed_position_cum, label = source)) +
geom_point(data = label_postions, color = "black",
aes(x = as_datetime(ymd(Date)-0.5),
y = fixed_position_cum, shape = source)) +
theme(legend.position = "right") +
#scale_color_nejm() +
scale_color_manual(values = col_scheme) +
guides(color = guide_legend(nrow = 2, byrow = TRUE))+
theme(axis.title.x = element_blank(), text = element_text(size = 16),
legend.position = "none") +
labs(color = "Source", shape = "Source",
y = "Cummulative number of deaths",
title = "Delays in the reported numbers of COVID-19 deaths in England and Wales")
discrepancies
# (discrepancies + theme(legend.position = "none") + labs(title = "[linear scale]")) /
# (discrepancies + scale_y_log10() + labs(title = "[log scale]")) +
# plot_annotation(title = "Discrepancies between the reported numbers")
Looking at the daily death rates it becomes clear that (1) data is very noisy and (2) there is a significant lag in the data.
We see that ONS data is especially influenced by the weekends and bank holidays. The reported numbers drop on the weekends (March 21 & 22, 28 & 29, April 4 & 5, Easter weekend, and so on) and are higher in the next days.
actual_vs_reported %>%
filter(source %in% names(col_scheme)) %>%
group_by(source) %>%
arrange(Date) %>%
mutate(deaths_on_that_day = number_of_deaths - lag(number_of_deaths)) %>%
ggplot(aes(x = Date, y = deaths_on_that_day, color = source, shape = source)) +
geom_smooth(se = FALSE, span = 0.3) +
geom_point(color = "black") +
geom_line(aes(group = source), alpha = 0.7, linetype = "dotdash") +
geom_text(data = label_postions, size = 7, hjust = 0,
aes(x = Date, y = fixed_position_daily, label = source)) +
geom_point(data = label_postions, color = "black",
aes(x = as_datetime(ymd(Date)-0.5),
y = fixed_position_daily, shape = source)) +
scale_color_manual(values = col_scheme) +
guides(color = guide_legend(nrow = 2, byrow = TRUE))+
labs(color = "Source", shape = "Source",
y = "Number of deaths on the day",
title = "Discrepancies between the reported numbers of COVID-19 deaths in England and Wales") +
theme(axis.title.x = element_blank(), legend.position = "none",
text = element_text(size = 16))
Age distribution
Looking at the deaths, Males are more affected than Females. How this plot looks like for cases? I.e. are men more susceptible?
I also wanted to compare the COVID-19 age and sex distribution with the UK population structure. I ended up comparing it to even more adequate data - to the deaths occuring in UK this year, before COVID-19. This will allow to put the age structure of COVID-19 into context.
max_axis <-
age_distribution %>%
group_by(sex, age_group) %>%
summarise(number_of_deaths = sum(number_of_deaths)) %>%
ungroup() %>%
filter(sex != "Both") %>%
pull(number_of_deaths) %>%
max()
max_axis <- round(max_axis / 100, 0) * 100 + 25
break_axis <- sort(c(-round(seq(0, max_axis, length.out = 5)),
round(seq(0, max_axis, length.out = 5))))
break_axis <- round(break_axis / 50) * 50
covid_age <-
age_distribution %>%
mutate(age_group = factor(age_group, levels = rev(age_levels))) %>%
filter(sex != "Both") %>%
mutate(nod = ifelse(sex == "Females", number_of_deaths, -number_of_deaths)) %>%
group_by(age_group) %>%
# filter(sum(number_of_deaths) > 0) %>%
ggplot(aes(x = age_group, y = nod, fill = sex)) +
geom_bar(stat = "identity") +
scale_fill_npg() +
scale_y_continuous(breaks = break_axis,
labels = abs(break_axis)) +
labs(x = "Age groups",
y = paste0("Number of deaths [data up to ", data_up_to, "]"),
fill = "Sex") +
coord_flip() +
theme(legend.position = "right")
before_covid <- ymd("2020-02-28")
age_distribution_bound <-
bind_rows(age_distribution_all %>%
mutate(type = "Before COVID-19"),
age_distribution %>%
mutate(type = "COVID-19")) %>%
filter(sex != "Both") %>%
group_by(sex, age_group, type) %>%
summarise(number_of_deaths = sum(number_of_deaths)) %>%
group_by(type) %>%
mutate(all_deaths = sum(number_of_deaths)) %>%
group_by(sex, type, age_group) %>%
mutate(fraction_of_deaths = number_of_deaths/all_deaths * 100) %>%
ungroup()
age_distribution_bound %>%
mutate(age_group = factor(age_group, levels = rev(age_levels)),
nod = ifelse(sex == "Females", number_of_deaths, -number_of_deaths)) %>%
group_by(age_group) %>%
filter(sum(number_of_deaths) > 0) %>%
ggplot(aes(x = age_group, y = nod, fill = sex)) +
geom_bar(stat = "identity") +
scale_fill_npg() +
facet_wrap(~type, scales = "free_x") +
# scale_y_continuous(breaks = break_axis,
# labels = abs(break_axis)) +
labs(x = "Age groups",
y = paste0("Number of deaths\n[Data up to:",
" before COVID-19 -- ", before_covid,
"; COVID-19 -- ", data_up_to,
"]"),
fill = "Sex",
title = ) +
coord_flip() +
theme(legend.position = "bottom",
text = element_text(size = 16))
This is even more visible when we compare the fraction of all deaths in each age group and with respect to sex.
age_distribution_bound %>%
group_by(age_group) %>%
filter(sum(fraction_of_deaths) > 5) %>%
ungroup() %>%
ggplot(aes(age_group, fraction_of_deaths, fill = type)) +
geom_bar(stat = "identity", position = "dodge") +
facet_wrap(~sex) +
scale_fill_jama() +
labs(title = "Fraction of all deaths with respect to age group and sex",
subtitle = "age groups contirbuting to fewer than 5% filtered out") +
theme(axis.title = element_blank(), legend.title = element_blank(),
text = element_text(size = 16))